This project is now in update mode. Check back regularly to see how things are progressing.
MetaSUB: Discover and Map the Genomes Around Us

Figure 1: Ubiquitous genomes are in, on, and around us. Source: Nature
Have you ever wondered what microscopic creatures you and your family come into contact with everyday? Our world is filled with trillions of microbes that you interact with everyday - those you breathe in, or get on your skin by touching other people or surfaces. Yet, we are only beginning to understand them.
DNA on any given surface you touch usually has 50-60% unknown species. Thus, there is still so much more to learn about the microbiome (all microscopic organisms) and the metagenome (all genomes their DNA) living in the world around us, affecting billions of hands every day, and knowing how these dynamics can affect and shape our health. But before we can learn about how microbiomes/metagenomes can impact humans (or how humans can impact them), we have first to learn exactly what is around us!
MAPPING THE WORLD'S METAGENOME
You are invited to support and contribute to an international research study about the urban metagenome as part of a global study on the genetic dynamics of our world’s cities (www.metasub.org).
In June of 2016 we launched the first-ever Global City Sampling Day (CSD), collecting DNA samples across the world’s cities, subways, and waterways. This international DNA and RNA collection effort is ongoing, through the hard work of trained medical students, public health officials, graduate students, engineers, designers, scientists, and other trained volunteers. The work in New York City is being done in collaboration with the Clinical and Translational Science Center (CTSC), CUNY Hunter College, Macaulay Honors College, City College, Cornell University, Columbia University, the Icahn School of Medicine at Mt. Sinai, Rockefeller University, and New York University.

Figure 2: Swabbing for discovery: MetaSUB Scientists (left to right) Sofia Ahsanuddin, Ebrahim Afshinnekoo, and Christopher Mason
The MetaSUB Consortium now works across 70 cities, scores of countries, and all seven continents around the world. We follow the Nagoya protocol in all countries that are signatories, and ensure complete transparency for our Consortium’s methods, goals, meetings, and results. All data are open and free for all to use.
Notably, the MetaSUB International Consortium also coordinates with the Global Ocean Sampling Day (OSD) group (https://www.microb3.eu/osd), to mirror our genetic snapshots of the world. Our Consortium efforts are funded by the Grand Challenges Explorations, an initiative of Bill and Melinda Gates Foundation, the United States National Institutes of Health (NIH), the WorldQuant Foundation, the Vallee Foundation, and is highlighted as part of the National Microbiome Initiative (NMI) of the United States.

Figure 3: Samples collected during Global City Sampling Day (CSD) 2016 and the Olympics in Rio (from http://metasub.org/interactive-map/).
HOW DOES IT WORK & WITH WHOM DOES IT WORK?
This research uses a nylon, sterile swab to collect samples from a surface in any surface or site. These samples will then be extracted for DNA/RNA, shotgun sequenced, and analyzed. We are investigating the microscopic and genetic world we interact with every day and that is shared between millions of people every day. The main objective is to map the DNA dynamics across the world’s cities and examine their signatures as they change from year-to-year, as well as during global human migration events (such as our study on the Olympics in 2016 and 2020).
This study has three specific aims, to: (1) globalize metagenome discovery and education, (2) track the density and dynamics of anti-microbial resistance (AMR) markers and biosynthetic gene clusters (BCGs), and (3) develop, optimize, and release open-source computational algorithms and tools for metagenomics research.
Protocols will follow standards from the International Metagenomics and Microbiome Standards Alliance (IMMSA), the MetaSUB Consortium, the F.D.A.’s Sequencing Quality Control Consortium, and optimized protocols from the fantastic Earth Microbiome Project (EMP) and the exciting eXtreme Microbiome Project (XMP). We also work with studies on extreme urban sites like the Gowanus Canal, in a project called the Brooklyn Bioreactor, and also the dynamics of urban bee hives in the Holobiont Urbanism project with the Playful Systems (Slavin) Lab at MIT.
HOW TO JOIN:
Get a hands-on experience by contributing samples from your home or city! Donors to this project will be sent a swab kit automatically.
WHERE DOES THE MONEY GO?
Even though the cost of sequencing is plummeting, it still is expensive to do for thousands of samples! We will use the money from this campaign to fund the processing of samples, expand our research, provide educational resources, and give students and post-docs fellowships to work on the project. The money will directly help the educational outreach, laboratory reagents, and the labor required to extract, prep, and sequence the DNA/RNA. To date, the MetaSUB project has largely been driven by high school, undergraduate, and graduate students working on a voluntary basis. Your money will generate stipends to support them as they continue to pursue their interests in science and research, notably as part of our work with Jeanne Garbarino in the Summer Science Research Program (SSRP) and the Community Research Education and Engagement for Data Science (CREEDS) Program at the Icahn School of Medicine at Mount Sinai.

Figure 4: Students and excited metagenomics researchers from the SSRP at Rockefeller University. Get your swab on.
THE WORKFLOW
This work is both time and resource-intensive. And we need more resources to advance the project's objectives and bold vision!
This is where YOU - The Citizen Scientist - come in! Now for the first time you’ll have the opportunity to discover what lives in your own home and community. By participating in our study, you can now understand what is on your toothbrush, your car, your home, your kid's lunchbox, or anything else you're curious about in your city!
 Figure 5: Collection Protocol for MetaSUB. Samples are collected (left), sequenced by next-generation methods (top right), and then analyzed on our large computing cluster (lower right) at the Institute for Computational Biomedicine (ICB) at Weill Cornell Medicine.
Figure 5: Collection Protocol for MetaSUB. Samples are collected (left), sequenced by next-generation methods (top right), and then analyzed on our large computing cluster (lower right) at the Institute for Computational Biomedicine (ICB) at Weill Cornell Medicine.
HOW METASUB WORKS
Becoming a Citizen Scientist is easy.
(1) Easy-to-use swabs for surface sampling. As pictured below, each swab will have a unique ID number that will be used to track the progress of your samples.
(2) Fool-proof instructions to ensure your samples yield enough material to enable accurate analyses of their DNA levels and types.
(3) A pre-stamped return envelope to ensure your swabs are quickly directed to our lab's facilities.
RESULTS - WHAT DO I GET?
For those getting digital reports on their samples, the Weill Cornell team will put together a comprehensive and interactive report that will not only tell you about your sample but allow you to explore it yourself!
The report will include (1) what microorganisms (taxa and species) discovered in your sample (2) what these organisms are doing (functional biochemical pathways) (3) the presence of any antimicrobial resistance markers (4) eukaryotes present - any plant, animal or fungi DNA (5) comparison to other databases - see what sample you closely resemble, are you a subway station or a water sample? (6) any novel biosynthetic gene clusters found - help expand the biochemical horizons of the science one swab at a time!
You now have the chance to explore this invisible world around you that you interact on a daily basis, what will you swab?
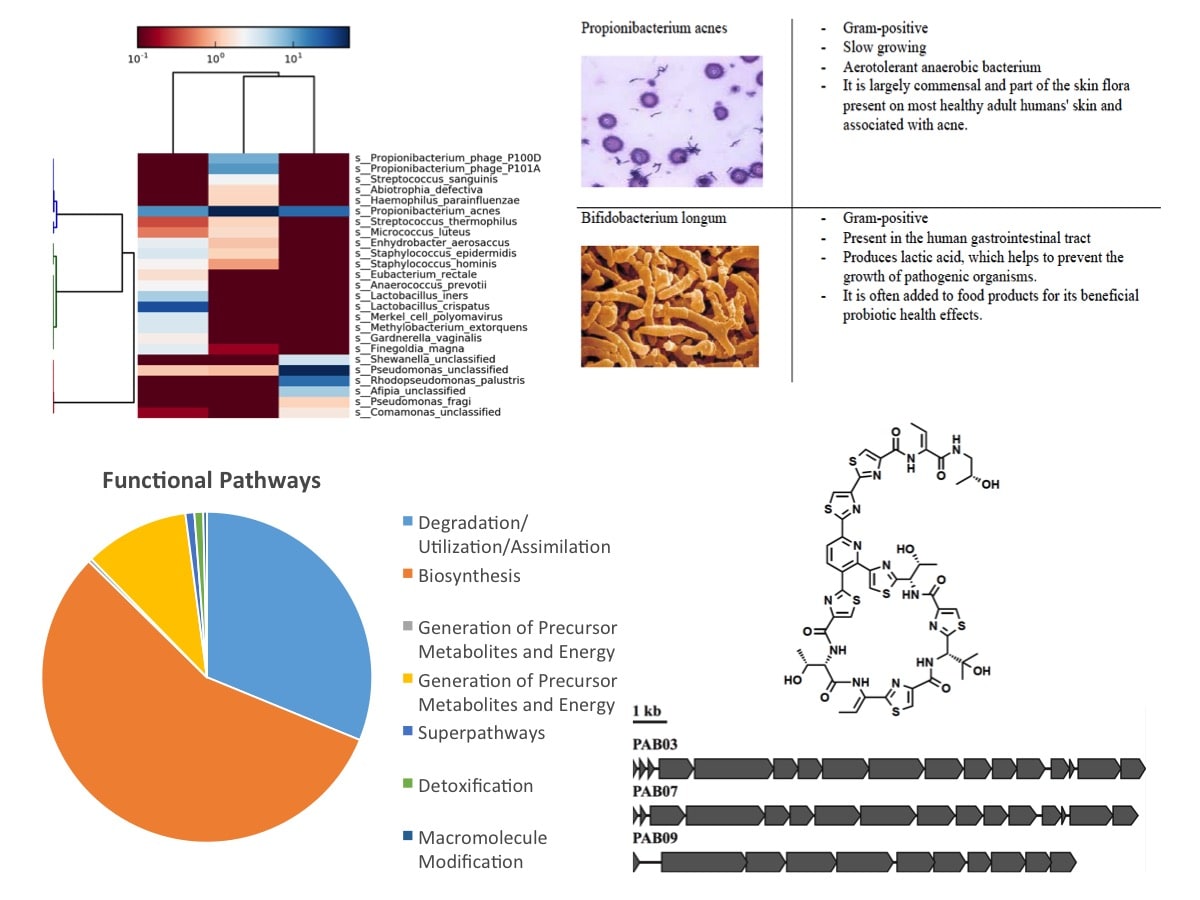
Figure 6: Annotation, Functional Classification, and Analysis Steps for MetaSUB samples. Thanks to Alexa McIntyre (Weill Cornell), Elizabeth Henaff (Weil Cornell), Peter Gonzalez (Weill Cornell), Stephen Woloszynek (Weill Cornell), and Mohamed Donia (Princeton University).
MATCHING FUNDS - How do I take advantage of my employer's matching gifts program?
When you file for the matching gift, include the fund number 0013619. The matching gift will be added to the project total when it is received.
THANK YOU!
We want to thank all of our funders listed above, all MetaSUB laboratories and scientists, and support from our industry partners that collaborate and contribute to our efforts, including Copan, CosmosID, GISCloud, IBM, Illumina, OneCodex, Promega, QIAGEN, and Zymo Research. Most importantly, we thank you!
For more details about the study, visit metasub.org and you can view articles from:











